| Mean centering |
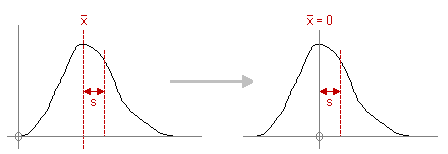 The selected data items (layers, spectra, or all data) are scaled in such a way that the mean of each item becomes zero. The selected data items (layers, spectra, or all data) are scaled in such a way that the mean of each item becomes zero. |
| Standardisation |
 The selected data items are scaled to a zero mean and a standard deviation of 1.0. Please note that the standardisation of data may be problematic if the data contains unusually large outliers. In this case q-normalisation is a better alternative. Note: the standardisation applied to spectra is also called "standard normal variate" (SNV). The selected data items are scaled to a zero mean and a standard deviation of 1.0. Please note that the standardisation of data may be problematic if the data contains unusually large outliers. In this case q-normalisation is a better alternative. Note: the standardisation applied to spectra is also called "standard normal variate" (SNV).
|
| Constant sum |
The selected items are scaled to a constant sum defined by the parameter A. |
| Constant sum of squares |
The selected items are scaled to a constant sum of squares. The sum is specified by the parameter A. |
| Maximum amplitude |
The selected items are scaled in such a way that the maximum absolute value of each item becomes A. |
| Range |
 The selected data values are scaled to cover a range between A and B. The selected data values are scaled to cover a range between A and B. |
| Q Normalisation |
 The selected data range is scaled to zero median and a difference between the median and the q-percentile of 1.0, with q (in %) given by the parameter A. Q normalisation is largely insensitive to outliers and should be used whenever you are expecting severe outliers. The selected data range is scaled to zero median and a difference between the median and the q-percentile of 1.0, with q (in %) given by the parameter A. Q normalisation is largely insensitive to outliers and should be used whenever you are expecting severe outliers. |
| Squashing Function |
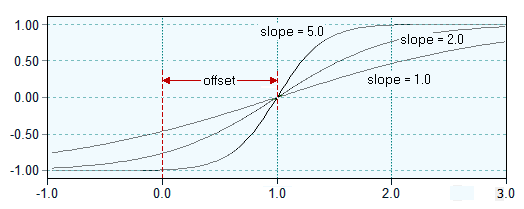 The selected data range is compressed by applying a sigmoid function ("squashing function") to the interval [-1,+1]. The parameter A specifies the origin (offset) of the squashing function, the parameter B defines the slope of the function. The selected data range is compressed by applying a sigmoid function ("squashing function") to the interval [-1,+1]. The parameter A specifies the origin (offset) of the squashing function, the parameter B defines the slope of the function.
|

 Preprocessing
Preprocessing  Scaling the Data
Scaling the Data


 Preprocessing > Scaling the Data
Preprocessing > Scaling the Data
 The selected data items (layers, spectra, or all data) are scaled in such a way that the mean of each item becomes zero.
The selected data items (layers, spectra, or all data) are scaled in such a way that the mean of each item becomes zero. The selected data items are scaled to a zero mean and a standard deviation of 1.0. Please note that the standardisation of data may be problematic if the data contains unusually large outliers. In this case q-normalisation is a better alternative. Note: the standardisation applied to spectra is also called "standard normal variate" (SNV).
The selected data items are scaled to a zero mean and a standard deviation of 1.0. Please note that the standardisation of data may be problematic if the data contains unusually large outliers. In this case q-normalisation is a better alternative. Note: the standardisation applied to spectra is also called "standard normal variate" (SNV).
 The selected data values are scaled to cover a range between A and B.
The selected data values are scaled to cover a range between A and B. The selected data range is scaled to zero median and a difference between the median and the q-percentile of 1.0, with q (in %) given by the parameter A. Q normalisation is largely insensitive to outliers and should be used whenever you are expecting severe outliers.
The selected data range is scaled to zero median and a difference between the median and the q-percentile of 1.0, with q (in %) given by the parameter A. Q normalisation is largely insensitive to outliers and should be used whenever you are expecting severe outliers. The selected data range is compressed by applying a sigmoid function ("squashing function") to the interval [-1,+1]. The parameter A specifies the origin (offset) of the squashing function, the parameter B defines the slope of the function.
The selected data range is compressed by applying a sigmoid function ("squashing function") to the interval [-1,+1]. The parameter A specifies the origin (offset) of the squashing function, the parameter B defines the slope of the function.